New research is helping to unveil how the parasite that causes sleeping sickness uses stealth tactics to escape detection by the human immune system.
New research is helping to unveil how the parasite that causes sleeping sickness uses stealth tactics to escape detection by the human immune system.
This may finally be one route to penetrating the stealth cloak of Trypanosoma brucei
Professor Mark Field
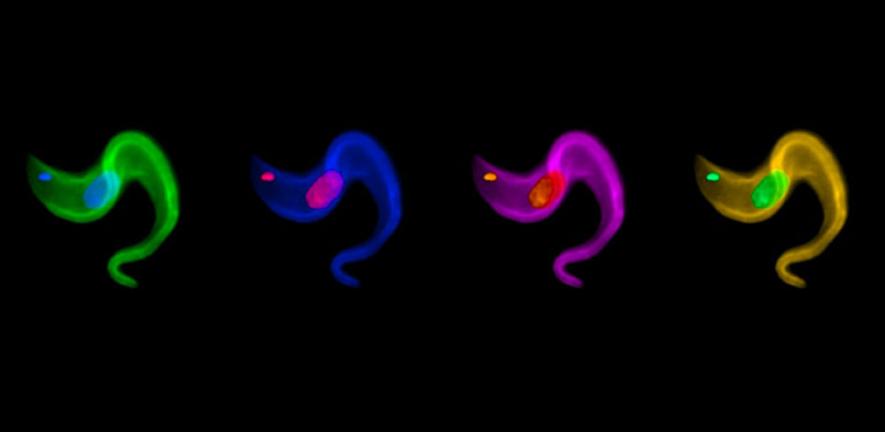
Stealth is a well-known concept in military tactics. Almost since the invention of radar, the hunt began for counter-technologies to hide aircraft and missiles from detection – most successfully by modifying the composition and shape of surfaces to confound detection. In a biological parallel, the African sleeping sickness parasite Trypanosoma brucei also has a stealth-like trick for altering its surface to confound recognition by the human immune system.
Trypanosomes are covered in some 10 million identical proteins called variant surface glycoproteins (VSGs). Although VSGs are well recognised by the immune system, trypanosomes can rapidly and repeatedly change every single one of these coat proteins, just as the rumblings of an immune response against the first coat have begun. In this way, the parasite can effectively ‘disappear’ again and again, deflecting the immune response to something that has essentially become an echo.
A complex mechanism of gene control underlies the process that allows trypanosomes to switch coats. Part of this mechanism has now been revealed in recent research by Cambridge parasitologist Professor Mark Field and colleagues from the USA, UK and Europe.
The finding has implications for treating African sleeping sickness, one of the ‘neglected tropical diseases’ – those that are caused by infectious agents that are endemic to low-income populations in Africa, Asia and the Americas, where treatment may not be universally available or is poor. Sleeping sickness threatens millions of lives in sub-Saharan countries and, because it also affects cattle, is a major contributor to economic hardship. Currently there is no vaccine.
At the heart of T. brucei’s switching mechanism is a set of genes, possibly as many as 2,000, encoding the VSGs. Only a single VSG gene is active at any one time – ensuring that the coat contains only a single protein. All other VSG genes are inactive or silent.
“The active VSG gene is within a site lying close to the end of the chromosome, while all the silent genes are elsewhere in the parasite genome,” said Field. “When switching occurs, the genes become rearranged and a gene from the silent archive is moved into the expression site. When this one is expressed, new coat proteins are moved to the cell surface to replace the old coat.”
The researchers have discovered a protein, NUP-1, which helps to maintain the silent archive, as Field described: “The basis of gene silencing is epigenetic. In other words, it’s not written in the DNA per se but is the result of protein ‘decorations’ called chromatin that either block or allow gene expression. NUP-1 is one of the proteins controlling epigenetic gene silencing.”
In multicellular organisms, a key regulator of chromatin remodelling is a protein called lamin, which forms a network across the inner lining of the cell’s nucleus. Until the discovery of NUP-1, an equivalent protein was unknown in plants, fungi or single-celled organisms. NUP-1 not only performs lamin-like functions – suggesting that the mechanism NUP-1 controls is probably an ancient process shared by organisms as diverse as humans and their parasites – but, excitingly, its discovery also opens up new possibilities for eradicating T. brucei by controlling coat switching.
“Without NUP-1, silenced genes are no longer constrained, and increased VSG switching happens,” explained Field. “If we could inactivate NUP-1, the parasite will probably exhaust the VSG repertoire or damage the surface coat so that it’s no longer effective as a barrier. This may finally be one route to penetrating the stealth cloak of Trypanosoma brucei.”
For more information, please contact Louise Walsh at the University of Cambridge Office of External Affairs and Communications.
This work is licensed under a Creative Commons Licence. If you use this content on your site please link back to this page.