Track and trace in Sierra Leone
How the genomics techniques keeping us safe from COVID-19 helped end the Ebola epidemic

Impact at a glance
- Ian Goodfellow helped set up the Ebola Treatment Centre in northern Sierra Leone and the Ebola Outbreak Sequencing Service, the country’s only genetic sequencing facility.
- He worked with the University of Makeni to establish a permanent Infectious Disease Research Laboratory.
- His team’s work directly informed the WHO’s Ebola response strategy.
- His contribution “formed a crucial role in helping to bring the epidemic in Sierra Leone to a close”.
- Their work helped inform technology used in the majority of SARS-CoV-2 sequencing.
"Professor Goodfellow’s contribution formed a crucial role in helping to bring the epidemic in Sierra Leone to a close and his ongoing work at the UniMak IDRL has allowed us to maintain effective local disease surveillance since."

Responding to a major outbreak
At the start of 2020, as it became apparent that the new respiratory disease first identified in China was beginning to spread beyond the country’s borders and healthcare professionals braced themselves for the oncoming storm, the global scientific community realised it had an important role to play.
Among these was Professor Ian Goodfellow from Cambridge’s Department of Pathology. Goodfellow was an expert in RNA viruses – viruses just like SARS-CoV-2, which was now spreading almost unchecked around the globe. Working originally on norovirus, he had shown that genetic sequencing could be used to track viral spread in a hospital setting, where an infection could cause serious complications for vulnerable patients.
But as COVID-19 began to take hold, Goodfellow faced an unusual predicament: all of his sequencing equipment was in the Democratic Republic of the Congo (DRC). He had taken it there in December 2019 to help a taskforce coordinated by the national research institute (INRB) to fight the emerging Ebola epidemic in DRC. (Fortunately, he was able to borrow equipment from the Cambridge community – and, within 24 hours of asking, began sequencing the SARS-CoV-2 virus in his lab at Addenbrooke’s Hospital.)
Until SARS-CoV2, Ebola was arguably one of the most serious – and terrifying – viruses to spread widely in humans since HIV. It causes a haemorrhagic fever that results in people literally bleeding to death in a half of all cases.
Although there were a small number of cases outside Africa – for example, a Scottish nurse and aid worker contracted the virus while working in Sierra Leone and was only diagnosed positive upon her return to the UK – most of the world escaped unscathed. In West Africa, however, the Ebola epidemic was devastating: more than 28,500 cases were recorded and 11,000 patients died. Countries including Guinea, Liberia and Sierra Leone united to try to contain the disease, but a lack of laboratory infrastructure and slow diagnostics hampered their initial efforts, allowing the epidemic to spread at an unprecedented rate.
Ebola virus particle (NIAID)
Ebola virus particle (NIAID)
In late 2014, Goodfellow headed to Sierra Leone to lend his support. Over the course of the epidemic, many more members of the Cambridge biomedical community – from the University and the diagnostic labs at the hospital – would join him, coordinated by Public Health England.
“I could see that the epidemic was getting out of control and that there were individuals like myself and my colleagues, molecular virologists, who had skills that would be useful, particular around diagnostics,” he says. “When it comes to Ebola, you need to identify people that are potentially infected, test them as quickly as possible and get them isolated if they’re positive.”
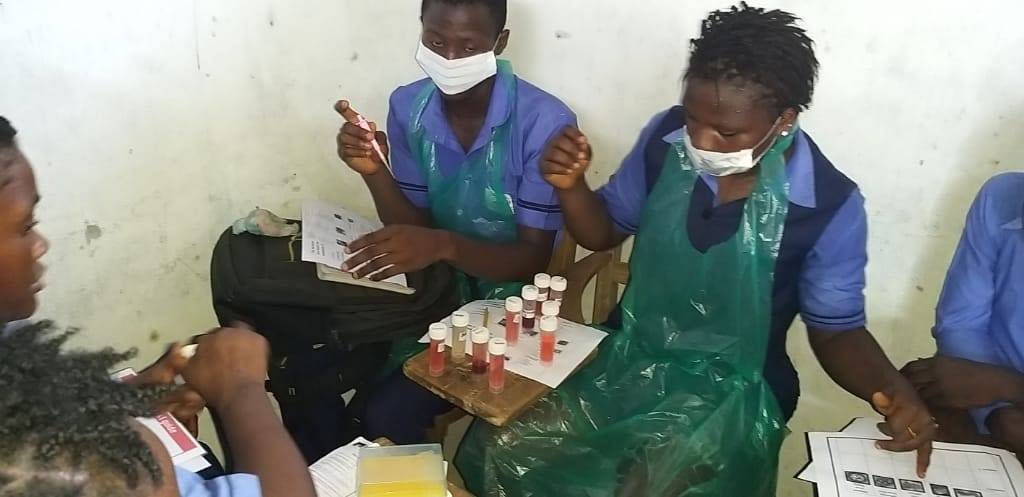
Goodfellow helped set up one of the first diagnostic laboratories in an Ebola Treatment Centre near Makeni, in northern Sierra Leone, with support from the UK government. He also established the Ebola Outbreak Sequencing Service, the country’s only genetic sequencing facility.
“You quickly realise the small things you take for granted,” he says. “So if you've got the wrong type of tubes and they don't fit in the centrifuge, for example, then you can't run your diagnostics. We didn't have a calibrated thermometer, so had no way of confirming that the temperatures required for heat inactivation of the samples were accurate. This meant in the first few months we had to do all the RNA extractions manually, which was a laborious, slow process.”
Poor internet capability meant he ended up sending results via his phone. “I ended up with a phone bill of £3,500!” he says.
Dr Armando Arias
Dr Armando Arias
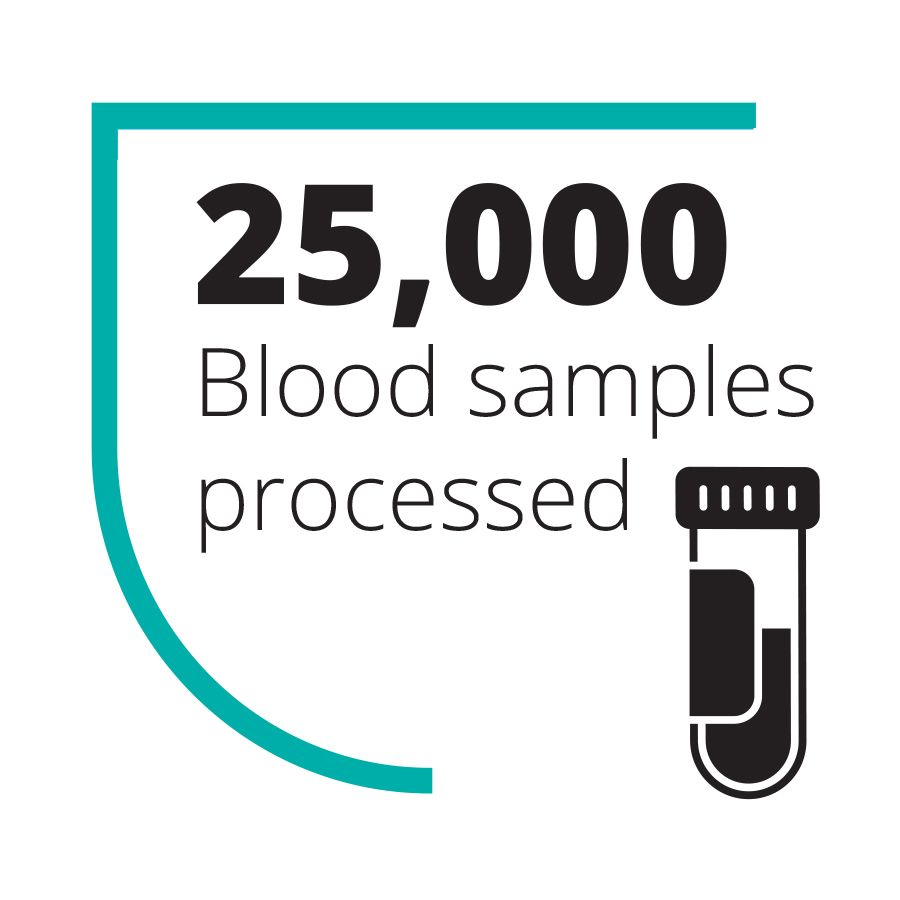
Over the course of the epidemic, the UK teams sent to work in the diagnostic lab that Goodfellow helped set up processed over 25,000 blood samples in the field. They reduced diagnosis time from seven days to 4–6 hours – vital for enabling non-infected patients, who would ordinarily be co-quarantined and potentially infected by diseased patients, to be discharged. (As an added bonus, they also detected cases of unrelated but treatable diseases, including over 750 patients with malaria who could then be given the appropriate treatment.)

Their work directly informed the WHO’s response strategy and provided the scientific basis for its coordinated quarantine and treatment of the 325 patients whose samples tested positive for Ebola at the Makeni treatment centre. Of the patients admitted to the treatment centre where Goodfellow worked, 40 individuals survived their infections.
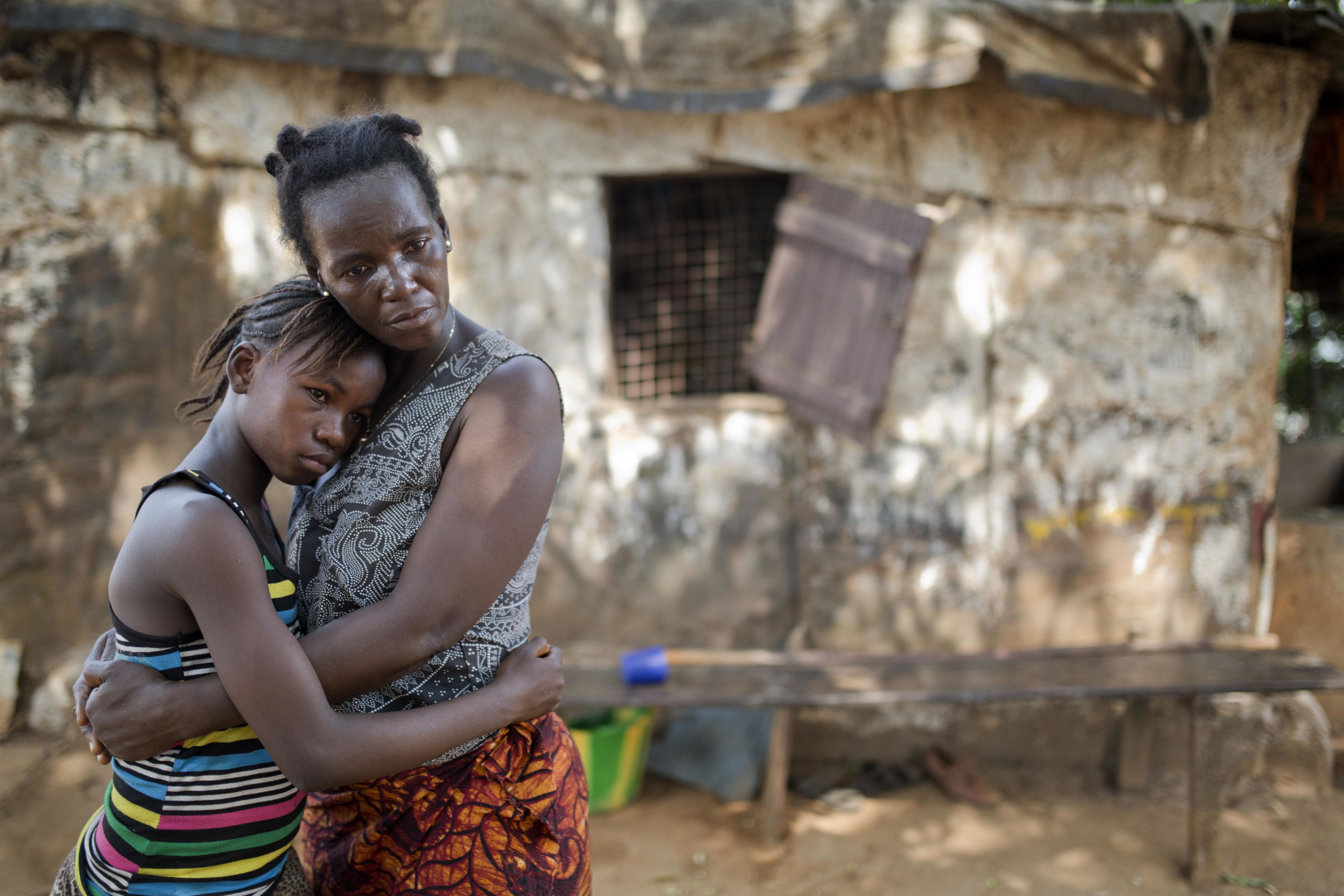
Building capacity
It was crucial to Goodfellow that they create a lasting legacy. “We needed to avoid ‘parachute science’, where scientists fly in, take samples and leave. It had to be about developing sustainable partnerships, about developing local capacity. With training and support, local researchers have the ability to respond to these outbreaks; they just need the equipment and the infrastructure.”
For this reason, Goodfellow worked with the University of Makeni to establish a permanent Infectious Disease Research Laboratory (UniMak-IDRL) within Sierra Leone, training local staff to provide diagnostics and state-of-the-art research methods. The laboratory continues to provide employment to local staff, supporting local public health efforts.

African researchers at the UniMak-IDRL
African researchers at the UniMak-IDRL
The strategy paid off. On 14 January 2016, following a 42-day period where no new cases had been identified, the WHO declared the Ebola epidemic in Sierra Leone over. That same day, with neither Goodfellow nor any of his UK colleagues in the country, a post-mortem sample tested positive: was there a new or previously-unmonitored chain of transmission? Was this single positive case just one of many undiagnosed cases in the community? The impact could have been devastating, requiring an immediate re-escalation of control measures.
Equipped to sequence the virus themselves within 48 hours, however, the laboratory staff were able to show that this was not a new or unmonitored outbreak. Rather, it was more likely that this case was linked to the re-emergence from a persistently-infected individual, possibly the individual’s sexual partner.
Similarly, when a 13-month old baby who had been quarantined for the standard 21 days developed the disease, the WHO considered increasing the quarantine period across the region, fearful that this case reflected an increase in the Ebola incubation period. This would have had major workforce, resource and containment implications for the response effort.
Fortunately, the team at the Ebola Outbreak Sequencing Service, working with colleagues from Birmingham, Edinburgh and the Wellcome Sanger Institute, was able to show that the infant had contracted the virus via breastmilk from an apparently healthy mother. Reassured by these results, the WHO maintained the existing 21-day quarantine period, and this research informed WHO guidelines for breastfeeding women in the context of Ebola.
"The UniMak Infectious Disease Research Laboratory and the training they have given staff, has provided much needed local expertise, allowing Sierra Leone to react quickly in response to any new, sporadic Ebola cases."
Combatting misinformation
As is the case increasingly with COVID-19, rumours and misinformation were rife during the Ebola epidemic, hampering efforts to control it. This prompted Goodfellow to establish a public engagement programme in 2016 to increase understanding of infectious diseases and how to protect oneself.
The programme currently employs three people and has engaged more than 4,000 students at over 50 schools across Sierra Leone. The team simulates real-time outbreaks and shows students how studying genetic sequences can help identify the source of the outbreak and inform control measures. Their work has continued during the COVID-19 pandemic, with the team continuing to engage local students with information on the pandemic and how to protect themselves and their families.











Blueprint for tackling COVID-19
Over the course of the epidemic and working alongside collaborators from the Wellcome Sanger Institute and the Universities of Edinburgh and Birmingham, Goodfellow’s team sequenced and analysed more than 1,200 samples from Ebola patients, generating over 600 complete genomes – more than a third of all the Ebola genomes produced during the epidemic.

Genomics surveillance of Ebola allowed scientists and public health officials to identify infection sources and clusters, implement ‘track and trace’ measures and provide public health teams with key data to contain and end the epidemic. If this sounds familiar, it is because it is the same approach that is used by the COVID-19 Genomics UK Consortium (COG-UK), set up by Professor Sharon Peacock at Cambridge, and of which Goodfellow is also a part.
“COG-UK builds on the expertise that comes from tackling outbreaks – from Sharon’s work on using genomic sequencing to tackle bacterial outbreaks in hospitals such as C. difficile, to our work and that of our collaborators on viral outbreaks such as Ebola in West Africa,” explains Goodfellow.
Goodfellow was one of the founders of the ARTIC Network, funded by Wellcome, which aims to develop real-time genomic surveillance in field settings. As well as delivering training workshops for researchers and staff from public health laboratories across Africa, the network has provided open source ‘blueprints’ to enable the setting up of diagnostic sequencing services in resource-limited settings. Already, this has helped in managing outbreaks including Ebola and rabies. Most recently its work is helping manage SARS-CoV-2 oubreaks.
“It’s difficult to get an absolute number, but we reckon as many as three-quarters of all COVID-19 sequences have been generated using ARTIC primer sets developed by my colleagues,” he says, referring to reagents used to amplify SARS-CoV-2 RNA to make it more easily readable, more cheaply.

Hand prints are from Ebola survivors. Each hand is a person who was admitted to the Ebola Treatment Centre and then discharged after they cleared Ebola.
Hand prints are from Ebola survivors. Each hand is a person who was admitted to the Ebola Treatment Centre and then discharged after they cleared Ebola.
Goodfellow is hugely grateful to the support he has received from the Wellcome Trust, who have funded him as a research fellow for almost twenty years, since 2003. He is currently a Wellcome Senior Fellow, which, he says, “gives me a lot of flexibility and protects my research time – it also enables me to be research active and work in the lab. This was critical to allowing me to set up the sequencing during Ebola – and now during COVID – as I was still able to work at the bench.”
Going into Sierra Leone at the start of the Ebola epidemic, Goodfellow admits to being “naïve” about the challenges of setting up sequencing facilities there and of convincing public health officials – who were already struggling to cope with the outbreak – of the value of sequencing. But the lessons he learnt there proved invaluable not only in helping end the epidemic in West Africa but in helping the world fight back against COVID-19.
"Ebola and COVID won’t be the last threats we face, but with each new outbreak we're getting better tooled up, better prepared to fight back and more able to use new advances in technology to inform public health responses."

With thanks to Richard Gilbertson, Anna Davies and Marwah Hassan for their contributions.
Images supplied by Ian Goodfellow, unless otherwise credited.
Infographics by Alison Fair