Hunting for COVID-19 variants
The story behind the UK's world-leading
SARS-CoV-2 genomics capability

For years, Professor Sharon Peacock has been using cutting-edge genome sequencing to track outbreaks of infectious diseases and to better understand how pathogens spread and evolve. When SARS-CoV-2 emerged just over a year ago, she knew that her work could play a vital role in helping the world fight back.
How scientists are helping us to understand how and why the COVID-19 virus is spreading, allowing us to fight back against the COVID-19 pandemic.
Towards the middle of December, a sombre Matt Hancock, UK Health Secretary, stood up to deliver an address to the House of Commons.
“Over the last few days, thanks to our world class genomic capability in the UK, we have identified a new variant of coronavirus, which may be associated with the faster spread in the south-east of England,” he told MPs.
It was the news that no one wanted to hear, yet which anyone who understood virus evolution might have anticipated. At the time, around 1,000 cases of infection with the new variant – dubbed the ‘Kent variant’ – had been identified. Just two months later, Professor Sharon Peacock, from Cambridge’s Department of Medicine, would tell the BBC that the variant had “swept the country", and was likely to spread worldwide.

Credit: Annie Spratt
Credit: Annie Spratt
If anyone was well placed to observe this sweep, it was Peacock. She leads the COVID-19 Genomics UK (COG-UK) Consortium, set up early on in the pandemic to deliver large-scale genome sequencing of SARS-CoV-2 to Public Health Agencies, the NHS and the UK government. COG-UK scientists together with Public Health England had been the first to realise that a variant first detected in Kent was rapidly establishing itself as the dominant variant in the UK.
This was not the first time that a variant had emerged and then spread rapidly.
“A variant first detected in Spain rapidly spread to many European countries over the summer months, and we were worried that it may be more transmissible,” explains Peacock, pulling up a slide that showed the SARS-CoV-2 lineages circulating in the UK since the start of the pandemic.
“If you look at the number of cases that we observed, there was a rapid surge in this variant in the UK – to the point where it became our most common circulating variant. But detailed analysis indicated that this pattern was most likely due to holiday travel.”
The Kent variant – known technically as B.1.1.7 – was different, however. Public health officials were seeing a surge of cases in Kent during a period of lockdown, while elsewhere in the UK numbers were stable. “That started to set alarm bells ringing,” she says.
When COG-UK scientists looked at the sequence data of this new variant, they saw that it had 23 differences in its genetic code compared to the original virus. Returning to the slide, Peacock points to a part of the virus ‘family tree’ that is based on comparing genomes with each other, pointing out what looks like an outlier, a distant, exotic relative – B.1.1.7. “It’s striking that it was clearly genetically different from other circulating SARS-CoV-2 in the UK.”
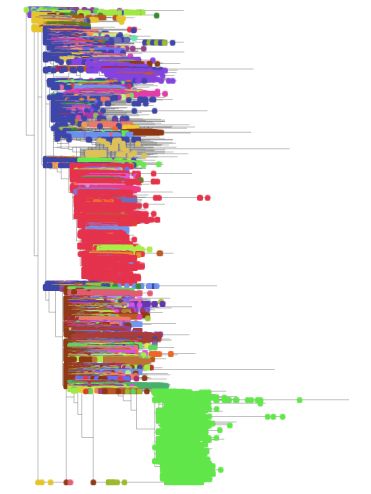
Phylogenetic tree showing SARS-CoV-2 lineages. B.1.1.7 is shown in green at the bottom.
Phylogenetic tree showing SARS-CoV-2 lineages. B.1.1.7 is shown in green at the bottom.
Looking back through their samples, they discovered that the first case of B.1.1.7 had appeared on 20 September 2020. For a while, there had been only a smattering of cases: it was only when this lineage began to expand towards the end of November and beginning of December that it became clear something was wrong.

Credit: Tim Dennell
Credit: Tim Dennell
Keeping an eye on the virus
Prior to the pandemic, Peacock had built up an international reputation as an expert in the use of genomics to track the spread of infectious diseases, including drug-resistant hospital-acquired infections such as MRSA and C. difficile.
Genomics is the study of an organism’s entire genetic code. The human genome is encoded as DNA, but for some viruses, including SARS-CoV-2, their genetic code is encoded as RNA, a series of nucleotides represented by the letters A, C, G and U.
Each time a virus replicates, errors can arise in its genetic code – ‘spelling mistakes’ that accumulate over time. SARS-CoV-2 is thought to mutate at a rate of around two nucleotides per month. Reading – or ‘sequencing’ – the genetic code of the virus can provide valuable information on its biology and transmission. It allows researchers to create phylogenetic trees – the ‘family trees’ that Peacock referred to earlier – which show how viruses are related to each other.
Early on during the pandemic, Peacock contacted a handful of experts in her field to canvass their thoughts on using sequencing to track the coronavirus. They were unanimous in their support, and from there, she says, everything snowballed.
On 11 March 2020, Peacock convened a meeting of 20 influential people in the field to thrash out how to make this work. “It was really that day that we hammered out the what, why, who and so on. That was on a Wednesday, and by the Sunday, we had a grant application on Sir Patrick Vallance’s desk [the government’s Chief Scientific Adviser]. The pace at which we moved was quite unusual.”

COG-UK officially came into life on 1 April 2020. It is led by Peacock and administered by the University of Cambridge. The Cambridge team manage the grant money, developed the legal, ethical and managerial framework, and run operations and logistics – it is “the heart of the operation”. But from the outset, Peacock has been keen to stress that the consortium is very much a collaboration, involving the four Public Health Agencies of the UK, numerous academic institutional partners, the Wellcome Sanger Institute, and diagnostic laboratory partners including the Lighthouse Labs and multiple NHS laboratories.
“When we first started, our ambition was to determine whether sequencing data could actually help inform public health actions and policy decisions. We all felt it was a bold step to take at the time. But actually, we've demonstrated that in spades.”
Why do mutations matter?
Viruses mutate all the time, and most mutations make little difference. But a small number will change the way the virus behaves. Peacock has a handy mnemonic to help her explain which mutations they are looking for: TILT.
T is for transmissible. If a mutation makes the virus more transmissible, it can spread more rapidly. The Kent variant spread across the UK in just a few months and is now seen in more than 90 countries.
I is for immunity. When our immune system encounters an infection, it triggers an immune response both to fight the current infection and to prevent future infection. Vaccines work by stimulating an immune response. If the virus mutates to evade this immune response, as has been seen in the South African variant, which partially evades the response, vaccines could become less effective.
L is for lethality. There is increasing evidence that the Kent variant can also cause more severe disease and potentially greater mortality.
T is for testing. Our ‘gold standard’ tests for COVID-19, known as PCR tests, could be altered by mutations in the genome. This is under regular review to ensure that tests do not become so affected that they report a false negative result.
“It’s really important to have eyes on the virus genome wherever you live, because the virus is evolving in every population all around the world. Genomics is an essential partner to vaccine effectiveness and where necessary, to their redesign to take account of variants of concern that avoid the immune response generated by natural infection or vaccination.”
SARS-CoV-2 (Credit: DataBase Center for Life Science)
SARS-CoV-2 (Credit: DataBase Center for Life Science)
World-leading expertise
Although COG-UK had been busy since day one, the consortium truly captured people’s attention with the emergence of the Kent variant. This, after all, was why COG-UK had been set up in the first place. Today, barely a day goes by without Peacock speaking to the media – both national and, increasingly, internationally.
While arranging one of these interviews, I had a chance to speak to a journalist in the USA who spoke rapturously about the consortium, astonished – almost angry – that her home nation seemed to be lagging way behind the UK when it came to genomic surveillance. As of mid-March 2021, COG-UK scientists have sequenced more than 350,000 virus samples, contributing just under half of all global SARS-CoV-2 sequence data deposited in GISAID, a major international open-access database.
When I ask why the UK is so far ahead, Peacock says she has puzzled over this for some time, but can only really say why she believes the UK is itself so strong (‘world-leading’, though she rarely uses this phrase herself). It is down to a combination of factors, she says.

Credit: Grooveland Designs
Credit: Grooveland Designs
Part of the reason is historic. The UK was an early adopter of using pathogen sequencing for public health. Every time a patient is diagnosed with TB, for example, their infecting organism is sequenced as a matter of routine, and the information is used to predict drug resistance and to look for TB outbreaks.
Then there is the matter of expertise. The UK has a large number of world experts in pathogen genomics, several of whom – such as Professor Ian Goodfellow at Cambridge – have worked on previous global emergencies, including the Ebola outbreak in West Africa. The UK also has strong networks of labs, including NHS labs, those of the four Public Health Agencies, and the Lighthouse Labs set up in response to SARS-CoV-2, as well as sequencing capability across regional labs and the Wellcome Sanger Institute.
Crucially, the information generated by these endeavours is shared, not squirrelled away for professional or commercial gain. “There's a philosophy that we give everything away. We give away all of our data, our methods and protocols. There are no egos in the room to stop that from happening.”
Despite being the Director of COG-UK and its driving force, Peacock feels uncomfortable when journalists give her too much credit. Referring to an article written in a major UK newspaper, she says, “I cringed when I read that Ravi [Gupta] and Ian [Goodfellow] were members of my group, since they are far more senior than this. They are independent PIs leading their own research teams.”
The autonomy of COG-UK researchers is important to the consortium’s success. “The way that the network was put together, it's a cooperative where we work together to contribute to the whole. [Our collaborators] work with us because they want to, and because it's important to the country. There's about 400 people in the network, and many of those people have volunteered.”
Much of what has been achieved has been done at no extra cost to the public purse. Nearly all of the COG-UK funds have been channelled into the costs of viral sequencing. Most of the collaborators are not paid by COG-UK funds, but instead are supported by their institutions. Rather than billing COG-UK for their researchers’ time and overheads, these institutions have essentially let their researchers work for free.
“Universities across the country have contributed hugely,” she says. “That's a staggeringly generous situation. [I want to] thank everybody, thank the universities, and thank people for their dedication. I think that universities don't really get enough of a shout out from all of this, but they have been utterly supportive of us.”

Credit: Tim Dennell
Credit: Tim Dennell
A lasting legacy
Faced with daily readouts of new infections and deaths, and news of emerging new variants, it’s sometimes hard to contemplate that we will – hopefully before too long – get through the pandemic. While people will argue about how the UK as a whole has responded to the pandemic – did we lock down too late, should we have eased restrictions sooner, for example – there are two groups who have received widespread recognition for their work: the NHS and the scientific community.
Cambridge Institute of Therapeutic Immunology & Infectious Disease (Credit: Lloyd Mann)
Cambridge Institute of Therapeutic Immunology & Infectious Disease (Credit: Lloyd Mann)
With its pioneering work on vaccines, clinical trials and now genomic surveillance, Peacock hopes the reputation of UK science will be a lasting legacy of the pandemic. She is heartened in particular to see female scientists come to the fore – Professor Sarah Gilbert at Oxford University, for example, can rightly take credit for helping the world emerge from the pandemic because of her vaccine research, but there are prominent female researchers at the head of several other vaccine development programmes.
“It’s important as a role model for girls and women in science to say, actually, you can be a leader in this, and you can stand up and be counted. That has to be a very loud and clear message that we send to people: that women can do science.”
Happy #InternationalWomensDay ♀️
— COVID-19 Genomics (COG-UK) Consortium (@CovidGenomicsUK) March 8, 2021
Today, we thank all women in the COG-UK network for your phenomenal efforts — be it in genome sequencing, technical expertise, logistics or scientific leadership. 👩🔬
Here are just some of the amazing women who have contributed to COG-UK 🏆 pic.twitter.com/IyBf0OufV8
And when we are finally able to return to some semblance of normality, what will become of COG-UK?
Peacock hopes that COG-UK will prove to be a forerunner of a UK-wide sequencing capability that can continue not just for SARS-CoV-2, but for other pathogens, too.
“The pace at which we've improved in sequencing – not just the technology itself, but the application of the technology – has advanced in a way that I would have never predicted. Let's look at how else we can use sequencing to improve our public health and in our clinical care. We have learned a really important lesson about the role of sequencing, and that will stand us in good stead if there's another pandemic.”
After all, she says, “Antimicrobial resistance is only around the corner, waiting for us.”

Credit: Tim Dennell
Credit: Tim Dennell